- Free and widely used open source database software
- Widely used in internet based applications
- Data are structured in
- Databases
- Tables within databases
- Fields within tables
- Each row is called a record
Reading mySQL
Jeffrey Leek
Johns Hopkins Bloomberg School of Public Health
mySQL
Example structure

Step 1 - Install MySQL

Step 2 - Install RMySQL
- On a Mac:
install.packages("RMySQL") - On Windows:
- Official instructions - http://biostat.mc.vanderbilt.edu/wiki/Main/RMySQL (may be useful for Mac/UNIX users as well)
- Potentially useful guide - http://www.ahschulz.de/2013/07/23/installing-rmysql-under-windows/
Example - UCSC database
UCSC MySQL
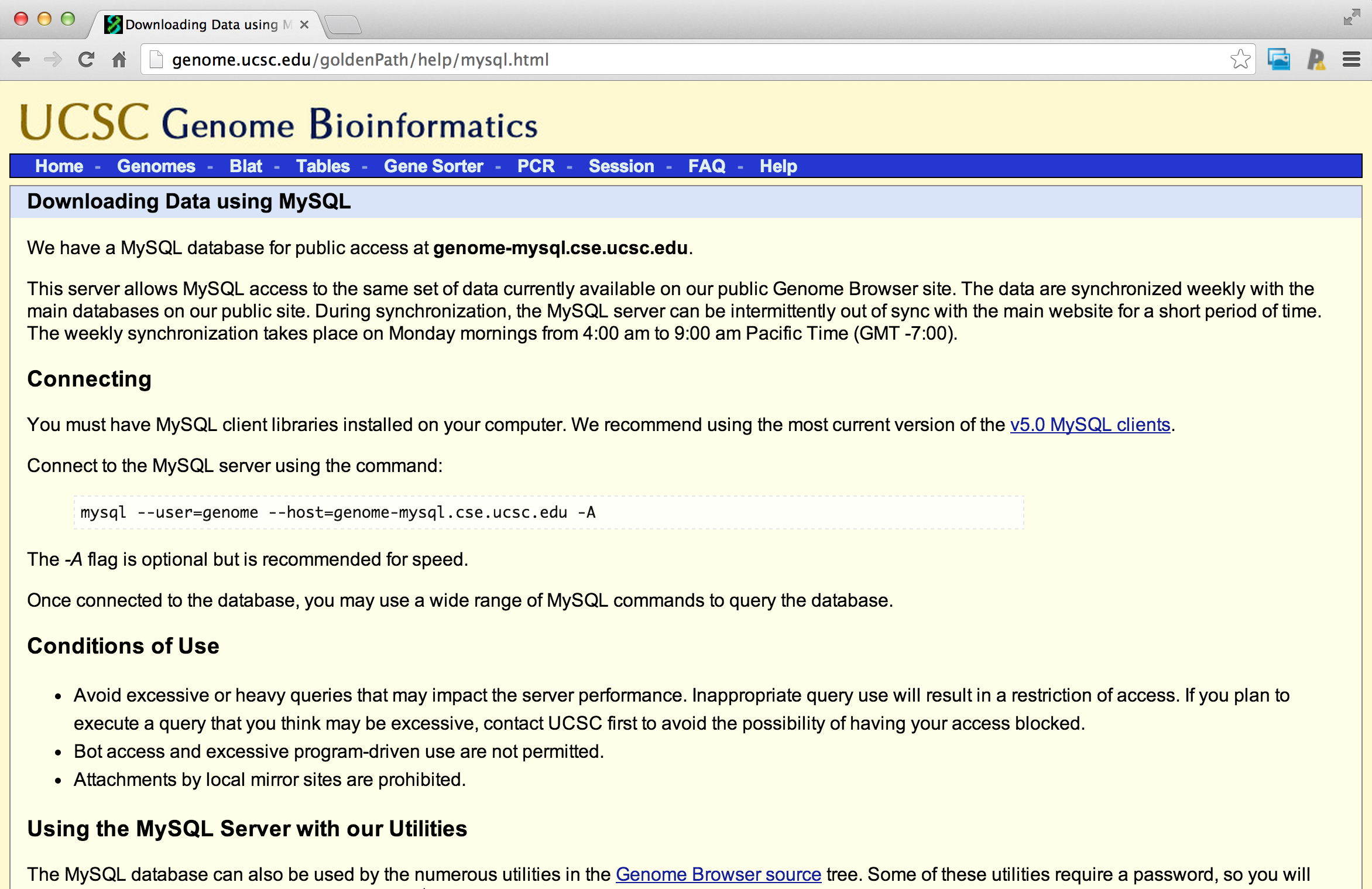
Connecting and listing databases
ucscDb <- dbConnect(MySQL(),user="genome",
host="genome-mysql.cse.ucsc.edu")
result <- dbGetQuery(ucscDb,"show databases;"); dbDisconnect(ucscDb);
[1] TRUE
result
Database
1 information_schema
2 ailMel1
3 allMis1
4 anoCar1
5 anoCar2
6 anoGam1
7 apiMel1
8 apiMel2
9 aplCal1
10 bosTau2
11 bosTau3
12 bosTau4
13 bosTau5
14 bosTau6
15 bosTau7
16 bosTauMd3
17 braFlo1
18 caeJap1
19 caePb1
20 caePb2
21 caeRem2
22 caeRem3
23 calJac1
24 calJac3
25 canFam1
26 canFam2
27 canFam3
28 cavPor3
29 cb1
30 cb3
31 ce10
32 ce2
33 ce4
34 ce6
35 cerSim1
36 choHof1
37 chrPic1
38 ci1
39 ci2
40 danRer1
41 danRer2
42 danRer3
43 danRer4
44 danRer5
45 danRer6
46 danRer7
47 dasNov3
48 dipOrd1
49 dm1
50 dm2
51 dm3
52 dp2
53 dp3
54 droAna1
55 droAna2
56 droEre1
57 droGri1
58 droMoj1
59 droMoj2
60 droPer1
61 droSec1
62 droSim1
63 droVir1
64 droVir2
65 droYak1
66 droYak2
67 echTel1
68 echTel2
69 equCab1
70 equCab2
71 eriEur1
72 felCat3
73 felCat4
74 felCat5
75 fr1
76 fr2
77 fr3
78 gadMor1
79 galGal2
80 galGal3
81 galGal4
82 gasAcu1
83 geoFor1
84 go
85 go080130
86 gorGor3
87 hetGla1
88 hetGla2
89 hg16
90 hg17
91 hg18
92 hg19
93 hg19Patch10
94 hg19Patch2
95 hg19Patch5
96 hg19Patch9
97 hgFixed
98 hgTemp
99 hgcentral
100 latCha1
101 loxAfr3
102 macEug1
103 macEug2
104 melGal1
105 melUnd1
106 micMur1
107 mm10
108 mm10Patch1
109 mm5
110 mm6
111 mm7
112 mm8
113 mm9
114 monDom1
115 monDom4
116 monDom5
117 musFur1
118 myoLuc2
119 nomLeu1
120 nomLeu2
121 nomLeu3
122 ochPri2
123 oreNil1
124 oreNil2
125 ornAna1
126 oryCun2
127 oryLat2
128 otoGar3
129 oviAri1
130 oviAri3
131 panTro1
132 panTro2
133 panTro3
134 panTro4
135 papAnu2
136 papHam1
137 performance_schema
138 petMar1
139 petMar2
140 ponAbe2
141 priPac1
142 proCap1
143 proteins120806
144 proteins121210
145 proteome
146 pteVam1
147 rheMac1
148 rheMac2
149 rheMac3
150 rn3
151 rn4
152 rn5
153 sacCer1
154 sacCer2
155 sacCer3
156 saiBol1
157 sarHar1
158 sorAra1
159 sp120323
160 sp121210
161 speTri2
162 strPur1
163 strPur2
164 susScr2
165 susScr3
166 taeGut1
167 tarSyr1
168 test
169 tetNig1
170 tetNig2
171 triMan1
172 tupBel1
173 turTru2
174 uniProt
175 vicPac1
176 vicPac2
177 visiGene
178 xenTro1
179 xenTro2
180 xenTro3
Connecting to hg19 and listing tables
hg19 <- dbConnect(MySQL(),user="genome", db="hg19",
host="genome-mysql.cse.ucsc.edu")
allTables <- dbListTables(hg19)
length(allTables)
[1] 10949
allTables[1:5]
[1] "HInv" "HInvGeneMrna" "acembly" "acemblyClass" "acemblyPep"
Get dimensions of a specific table
dbListFields(hg19,"affyU133Plus2")
[1] "bin" "matches" "misMatches" "repMatches" "nCount" "qNumInsert"
[7] "qBaseInsert" "tNumInsert" "tBaseInsert" "strand" "qName" "qSize"
[13] "qStart" "qEnd" "tName" "tSize" "tStart" "tEnd"
[19] "blockCount" "blockSizes" "qStarts" "tStarts"
dbGetQuery(hg19, "select count(*) from affyU133Plus2")
count(*)
1 58463
Read from the table
affyData <- dbReadTable(hg19, "affyU133Plus2")
head(affyData)
bin matches misMatches repMatches nCount qNumInsert qBaseInsert tNumInsert tBaseInsert strand
1 585 530 4 0 23 3 41 3 898 -
2 585 3355 17 0 109 9 67 9 11621 -
3 585 4156 14 0 83 16 18 2 93 -
4 585 4667 9 0 68 21 42 3 5743 -
5 585 5180 14 0 167 10 38 1 29 -
6 585 468 5 0 14 0 0 0 0 -
qName qSize qStart qEnd tName tSize tStart tEnd blockCount
1 225995_x_at 637 5 603 chr1 249250621 14361 15816 5
2 225035_x_at 3635 0 3548 chr1 249250621 14381 29483 17
3 226340_x_at 4318 3 4274 chr1 249250621 14399 18745 18
4 1557034_s_at 4834 48 4834 chr1 249250621 14406 24893 23
5 231811_at 5399 0 5399 chr1 249250621 19688 25078 11
6 236841_at 487 0 487 chr1 249250621 27542 28029 1
blockSizes
1 93,144,229,70,21,
2 73,375,71,165,303,360,198,661,201,1,260,250,74,73,98,155,163,
3 690,10,32,33,376,4,5,15,5,11,7,41,277,859,141,51,443,1253,
4 99,352,286,24,49,14,6,5,8,149,14,44,98,12,10,355,837,59,8,1500,133,624,58,
5 131,26,1300,6,4,11,4,7,358,3359,155,
6 487,
qStarts
1 34,132,278,541,611,
2 87,165,540,647,818,1123,1484,1682,2343,2545,2546,2808,3058,3133,3206,3317,3472,
3 44,735,746,779,813,1190,1195,1201,1217,1223,1235,1243,1285,1564,2423,2565,2617,3062,
4 0,99,452,739,764,814,829,836,842,851,1001,1016,1061,1160,1173,1184,1540,2381,2441,2450,3951,4103,4728,
5 0,132,159,1460,1467,1472,1484,1489,1497,1856,5244,
6 0,
tStarts
1 14361,14454,14599,14968,15795,
2 14381,14454,14969,15075,15240,15543,15903,16104,16853,17054,17232,17492,17914,17988,18267,24736,29320,
3 14399,15089,15099,15131,15164,15540,15544,15549,15564,15569,15580,15587,15628,15906,16857,16998,17049,17492,
4 14406,20227,20579,20865,20889,20938,20952,20958,20963,20971,21120,21134,21178,21276,21288,21298,21653,22492,22551,22559,24059,24211,24835,
5 19688,19819,19845,21145,21151,21155,21166,21170,21177,21535,24923,
6 27542,
Select a specific subset
query <- dbSendQuery(hg19, "select * from affyU133Plus2 where misMatches between 1 and 3")
affyMis <- fetch(query); quantile(affyMis$misMatches)
0% 25% 50% 75% 100%
1 1 2 2 3
affyMisSmall <- fetch(query,n=10); dbClearResult(query);
[1] TRUE
dim(affyMisSmall)
[1] 10 22
Don't forget to close the connection!
dbDisconnect(hg19)
[1] TRUE
Further resources
- RMySQL vignette http://cran.r-project.org/web/packages/RMySQL/RMySQL.pdf
- List of commands http://www.pantz.org/software/mysql/mysqlcommands.html
- Do not, do not, delete, add or join things from ensembl. Only select.
- In general be careful with mysql commands
- A nice blog post summarizing some other commands http://www.r-bloggers.com/mysql-and-r/