When you download R from the Comprehensive R Archive Network (CRAN), you get that ``base" R system
The base R system comes with basic functionality; implements the R language
One reason R is so useful is the large collection of packages that extend the basic functionality of R
R packages are developed and published by the larger R community
Installing R Packages
Jeffrey Leek
Johns Hopkins Bloomberg School of Public Health
R Packages
Obtaining R Packages
The primary location for obtaining R packages is CRAN
For biological applications, many packages are available from the Bioconductor Project
You can obtain information about the available packages on CRAN with the
available.packages()function
a <- available.packages()
head(rownames(a), 3) ## Show the names of the first few packages
## [1] "A3" "abc" "abcdeFBA"
There are approximately 5200 packages on CRAN covering a wide range of topics
A list of some topics is available through the Task Views link, which groups together many R packages related to a given topic
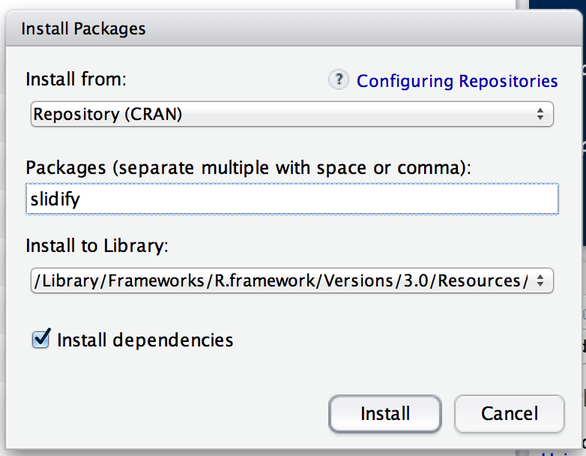
Installing an R Package
Packages can be installed with the
install.packages()function in RTo install a single package, pass the name of the lecture to the
install.packages()function as the first argumentThe following the code installs the slidify package from CRAN
install.packages("slidify")
This command downloads the slidify package from CRAN and installs it on your computer
Any packages on which this package depends will also be downloaded and installed
Installing an R Package
You can install multiple R packages at once with a single call to
install.packages()Place the names of the R packages in a character vector
install.packages(c("slidify", "ggplot2", "devtools"))
Installing an R Package in RStudio
Installing an R Package in RStudio
Installing an R Package from Bioconductor
- To get the basic installer and basic set of R packages (warning, will install multiple packages)
source("http://bioconductor.org/biocLite.R")
biocLite()
- Place the names of the R packages in a character vector
biocLite(c("GenomicFeatures", "AnnotationDbi"))
Loading R Packages
Installing a package does not make it immediately available to you in R; you must load the package
The
library()function is used to load packages into RThe following code is used to load the ggplot2 package into R
library(ggplot2)
Any packages that need to be loaded as dependencies will be loaded first, before the named package is loaded
NOTE: Do not put the package name in quotes!
Some packages produce messages when they are loaded (but some don't)
Loading R Packages
After loading a package, the functions exported by that package will
be attached to the top of the search() list (after the workspace)
library(ggplot2)
search()
## [1] ".GlobalEnv" "package:kernlab" "package:caret"
## [4] "package:lattice" "package:ggplot2" "package:makeslides"
## [7] "package:knitr" "package:slidify" "tools:rstudio"
## [10] "package:stats" "package:graphics" "package:grDevices"
## [13] "package:utils" "package:datasets" "package:methods"
## [16] "Autoloads" "package:base"
Summary
R packages provide a powerful mechanism for extending the functionality of R
R packages can be obtained from CRAN or other repositories
The
install.packages()can be used to install packages at the R consoleThe
library()function loads packages that have been installed so that you may access the functionality in the package